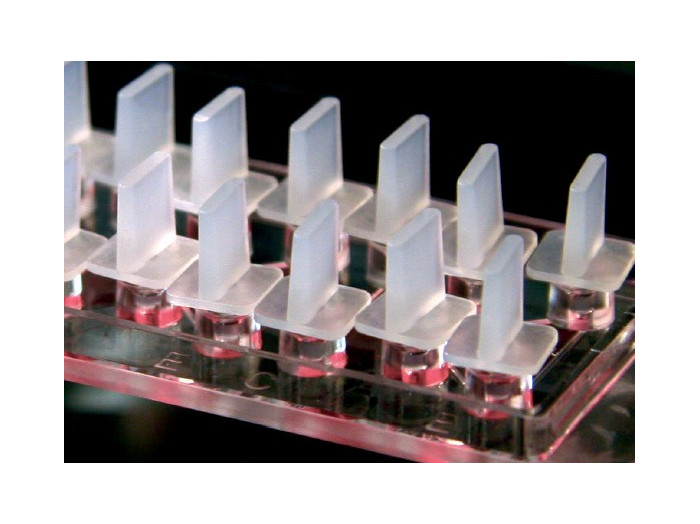
µ-Slide สำหรับการตรวจสอบ chemotaxis ที่มีการเคลื่อนที่ช้าหรือเร็วของเซลล์เกาะติด หรือเซลล์ลอย
- สามารถวัด chemotaxis แบบ real time ทั้งในสภาพแวดล้อมแบบ 2 มิติ หรือ 3 มิติ
- มีความสเถียรของ chemotactic gradients สำหรับการทดลองในระยะยาว
- สามารถทำการทดลองซ้ำได้ โดยมีความน่าเชื่อถือของผลการทดลอง
Investigate chemotaxis of fast or slow migrating adherent cells and non-adherent cells in 2D or 3D gel matrices
- Real-time chemotaxis measurement in a 2D or 3D environment
- Stable chemotactic gradients for long-term experiments
- Reproducible results with reliable and user-independent data
- Surface Modification: ibiTreat: #1.5 polymer coverslip, tissue culture treated, sterilized
Pcs./Box: 10 (individually packed) - Surface Modification: Collagen IV: #1.5 polymer coverslip, sterilized
Pcs./Box: 10 (individually packed)
Applications
- 2D and 3D chemotaxis assays with fast or slow migrating cells
- Live cell imaging using inverted microscopy
- Chemotaxis of neutrophiles, lymphocytes, and monocytes
- 3D chemotaxis of leukocytes or cancer cells in an ECM-like matrix
- Invasion assays of tumor cells in Matrigel™
- Chemotaxis measurements of adherent and non-adherent cells
Specifications
| Outer dimensions (w x l) | 25.5 x 75.5 mm² |
| Chemotaxis chambers on slide | 3 |
| Volume per chamber | 130 µl |
| Observation area | 2×1 mm² |
| Coating area per chamber – When coating full chamber – When coating observation area only | 3.50 cm2 0.27 cm2 |
| Distance between chambers | 18.5 mm |
| Total height with plugs | 12 mm |
| Volume chemoattractant | 30 µl |
| Bottom: ibidi Polymer Coverslip | |
Technical Features
- Chamber geometry optimized for cells in 2D and 3D matrices
- Defined linear gradient with long-term stability
- Homogeneous cell distribution at the experimental starting point
- 3 chambers on one slide for parallel assays
- Ideal for collagen gels, hydrogels, Matrigel™, or similar aqueous gels
- Ready to use, i.e., no assembly required
- Lettered and numbered chambers and reservoirs
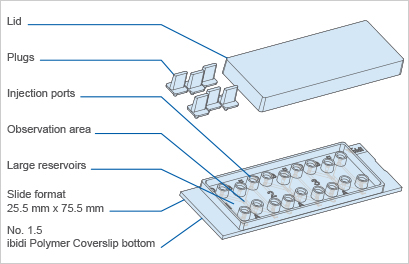
The Principle of the µ-Slide Chemotaxis
The μ-Slide Chemotaxis includes three chambers for three parallel assays. One chamber consists of two large reservoirs that are connected by a narrow observation area.
The µ-Slide Chemotaxis was developed to investigate the chemotactical behavior of fast or slow migrating, non-adherent or adherent cells on 2D surfaces or in 3D gel matrices. It is possible to observe the cell migration in linear and stable concentration profiles for more than 48 hours. As gradients are rapidly established, fast migration responses (occurring in less than 30 minutes) can also be measured.
Using the µ-Slide Chemotaxis allows for detailed and defined analysis of the migration behavior of various cell types, such as endothelial cells, fibroblasts, cancer cells, and immune cells.
Zengel P, et al. (2011) μ-Slide Chemotaxis: a new chamber for long-term chemotaxis studies. BMC Cell Biol 12:21. 10.1186/1471-2121-12-21.
read abstract
Biswenger V, et al. (2018) Characterization of EGF-guided MDA-MB-231 cell chemotaxis in vitro using a physiological and highly sensitive assay system. PLoS One 13(9):e0203040. 10.1371/journal.pone.0203040.
read abstract
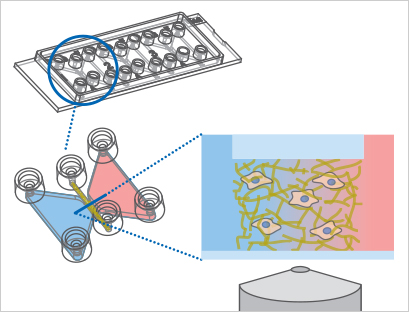
Schematic of the µ-Slide Chemotaxis principle. In this example, a 3D experiment with migratory cells in a gel matrix is shown.
Sample Preparation
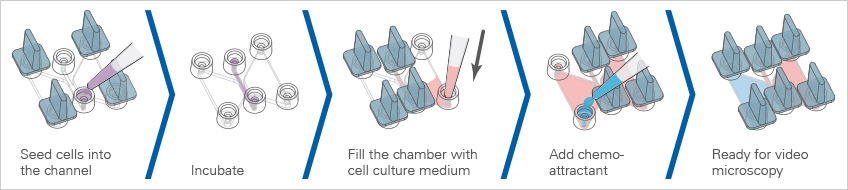
Experimental Workflow
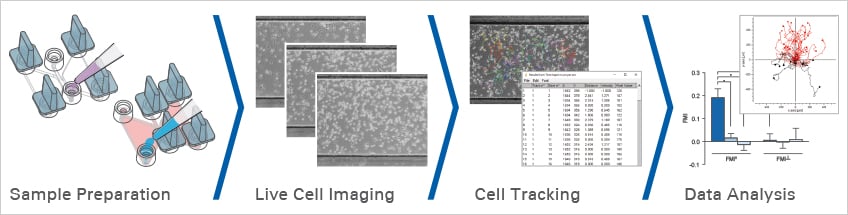
ibidi provides the complete solution for your chemotaxis experiment:
- Sample preparation using the µ-Slide Chemotaxis
- Live cell imaging with the ibidi Heating and Gas Incubation System
- Cell tracking using the free Manual Tracking plugin for ImageJ
- Data analysis with the free Chemotaxis and Migration Tool
- Fully automated image analysis with Chemotaxis FastTrack AI
Explore the detailed experimental workflow of a chemotaxis assay in our applications section.
ibidi Tutorial

Check out our online training for easy, fast, and accurate AI-based analysis of chemotaxis time-lapse movies here.
Example Data of a Chemotaxis Assay
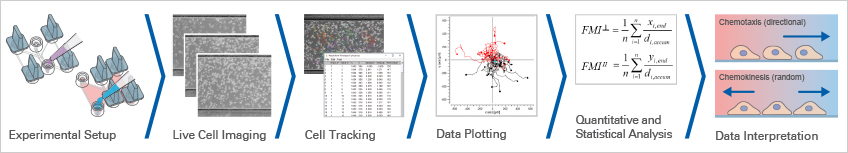
Here, we provide all files that were created during a chemotaxis experiment for download, to give you the ability to practice every step of the analysis and interpretation of chemotaxis data.
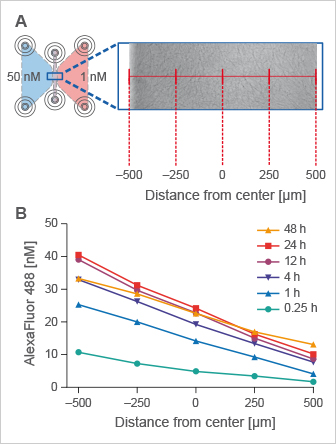
Gradient Stability in the µ-Slide Chemotaxis
The ibidi μ-Slide Chemotaxis is designed to provide a quick gradient with excellent long-term stability for more than 48 hours. The chamber design is ideally suited for chemotaxis assays with both fast migrating (e.g., leukocytes) and slow migrating cell types (e.g., cancer cells).
In 3D chemotaxis assays using aqueous gels, such as Collagen I or Matrigel, the gradient is stably established and not influenced by the gel in any way.
Time-stable gradients across the observation area in the µ-Slide Chemotaxis. (A) Top view of the chemotaxis chamber with a collagen I matrix in the observation area. The reservoirs were filled with 50 nM and 1 nM AlexaFluor 488, respectively. Measurements were performed at five different x-positions, indicated in red. (B) Time-stable linear AlexaFluor 488 concentration profiles were created within the migration chamber, measured by fluorescence correlation spectroscopy.
Application Examples Using the µ-Slide Chemotaxis
Live Cell Imaging of Actin Dynamics in a Chemotactic Gradient
F-actin networks play an important role during cell migration, which can be investigated in detail using chemotactic gradients. Primary dendritic cells were isolated from mice and transfected with the LifeAct Plasmid.
For the chemotaxis assay, cells were seeded on the µ-Slide Chemotaxis and a chemotactic gradient (CCL19) was applied. One day after the transfection, F-actin dynamics in the migrating cells were visualized using live cell imaging.
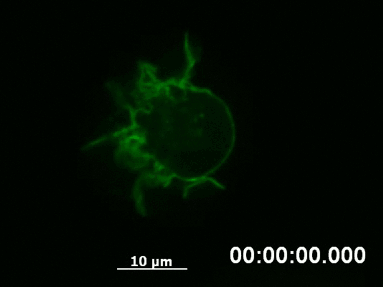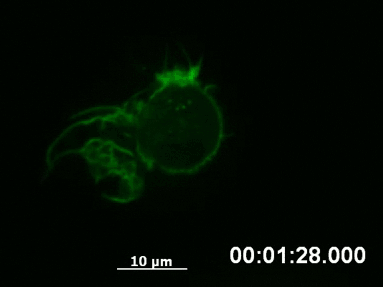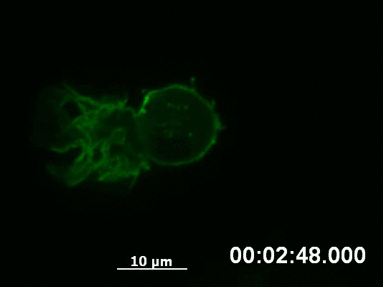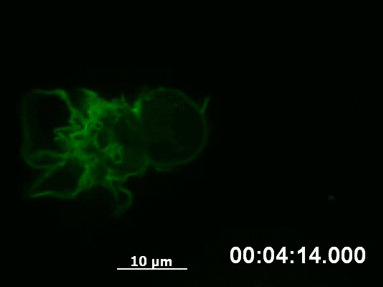
Live cell imaging of actin dynamics in a LifeAct-expressing primary dendritic mouse cell after the application of a chemotactic gradient.
Chemotaxis of Leukocytes in 3D
Fast leukocytes, such as dendritic cells or T-cells, can be visualized well during chemotaxis when inside gel matrices like collagen. This is especially true when using high-resolution fluorescence microscopy, which shows changes in the cytoskeleton during immune response and 3D interstitial migration.
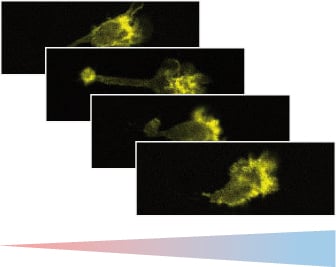
Fluorescence microscopy of murine chemotactic dendritic cells. The cytoskeleton is visualized using LifeAct.
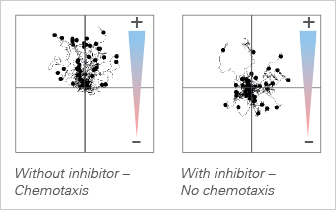
Chemotaxis Inhibition in Endothelial Cells (HUVEC)
Endothelial cells form the tip of the spear when blood vessels are generated during tumor development. Blocking this chemotaxis-driven mechanism is one of the key goals in anti-angiogenic therapeutic approaches. With the µ-Slide Chemotaxis, endothelial cells can be exposed to compounds that inhibit chemotaxis functionality.
Watch the movie: Chemotaxis of HUVEC Blocked by Spongistatin
Rothmeier AS, et al. (2009) Investigation of the marine compound spongistatin 1 links the inhibition of PKCα translocation to nonmitotic effects of tubulin antagonism in angiogenesis. FASEB J 23(4):1127–1137. 10.1096/fj.08-117127
read abstract
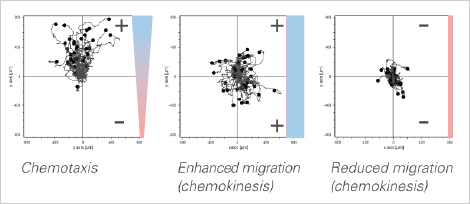
Distinguishing Chemotaxis and Chemokinesis in Cancer Cells
Chemotaxis, described as the directed motion towards a chemoattractant, can be distinguished from chemokinesis, which is known as an enhanced migration effect.
The µ-Slide Chemotaxis allows for the analysis of both effects, independent of each other (unlike transwell assays). In the example, the chosen chemoattractant induces both chemotaxis and chemokinesis of cancer cells.
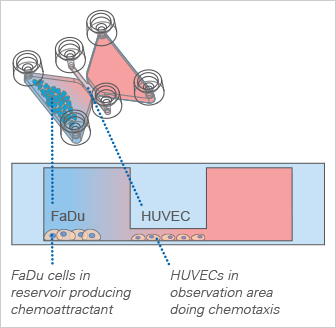
Cell-to-Cell Chemotaxis
The ibidi μ-Slide Chemotaxis can additionally be used for cell-to-cell chemotaxis assays. For example, cells brought inside the large reservoirs can be used as a chemoattractant producer.
In the example shown in the figure (Zengel et al., 2011), the cell line FaDu was chosen as a possible source for a chemoattractant. Endothelial cells (HUVEC) were tested for their chemotactic response. HUVEC show a similar chemotaxis effect in direction towards the FaDu cells, which is comparable to the chemotaxis effect towards 10% fetal calf serum as chemoattractant.
Zengel P, et al. (2011) μ-Slide Chemotaxis: a new chamber for long-term chemotaxis studies. BMC Cell Biol 12:21. 10.1186/1471-2121-12-21.
read abstract